18. UMAP图绘制
清除当前环境中的变量
rm(list=ls())
设置工作目录
setwd("C:/Users/Dell/Desktop/R_Plots/18umap/")
查看示例数据
head(iris)
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
iris.data = iris[,c(1:4)]
iris.labels = iris$Species
head(iris.data)
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## 1 5.1 3.5 1.4 0.2
## 2 4.9 3.0 1.4 0.2
## 3 4.7 3.2 1.3 0.2
## 4 4.6 3.1 1.5 0.2
## 5 5.0 3.6 1.4 0.2
## 6 5.4 3.9 1.7 0.4
head(iris.labels)
## [1] setosa setosa setosa setosa setosa setosa
## Levels: setosa versicolor virginica
使用umap包进行UMAP降维可视化分析
library(umap)
# 使用umap函数进行UMAP降维分析
iris.umap = umap::umap(iris.data)
iris.umap
## umap embedding of 150 items in 2 dimensions
## object components: layout, data, knn, config
# 查看降维后的结果
head(iris.umap$layout)
## [,1] [,2]
## [1,] 12.59949 3.759360
## [2,] 13.80894 5.272737
## [3,] 14.43500 4.813293
## [4,] 14.36174 4.896541
## [5,] 12.80543 3.473356
## [6,] 12.14902 2.715970
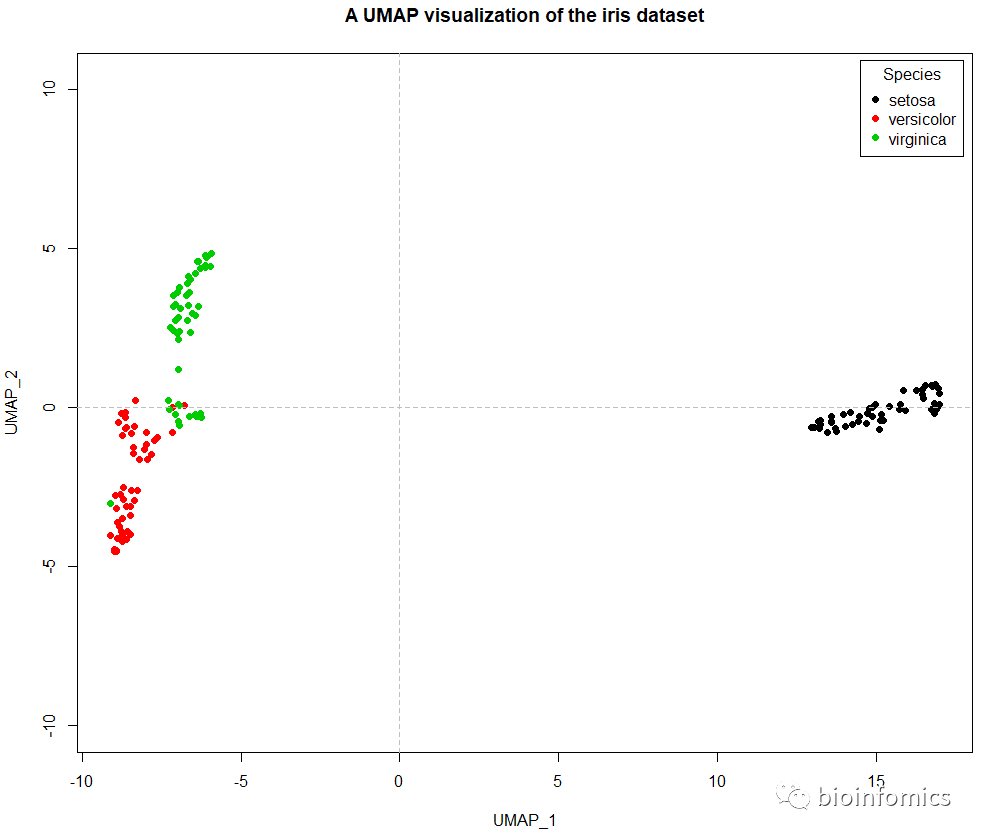
# 使用plot函数可视化UMAP的结果
plot(iris.umap$layout,col=iris.labels,pch=16,asp = 1,
xlab = "UMAP_1",ylab = "UMAP_2",
main = "A UMAP visualization of the iris dataset")
# 添加分隔线
abline(h=0,v=0,lty=2,col="gray")
# 添加图例
legend("topright",title = "Species",inset = 0.01,
legend = unique(iris.labels),pch=16,
col = unique(iris.labels))
使用uwot包进行UMAP降维可视化分析
library(uwot)
head(iris)
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
# 使用umap函数进行UMAP降维分析
iris_umap <- uwot::umap(iris)
head(iris_umap)
## [,1] [,2]
## [1,] -10.009518 -4.084256
## [2,] -9.505712 -5.754688
## [3,] -10.056058 -5.995699
## [4,] -9.884181 -6.000248
## [5,] -10.066767 -4.143123
## [6,] -10.620676 -3.369200
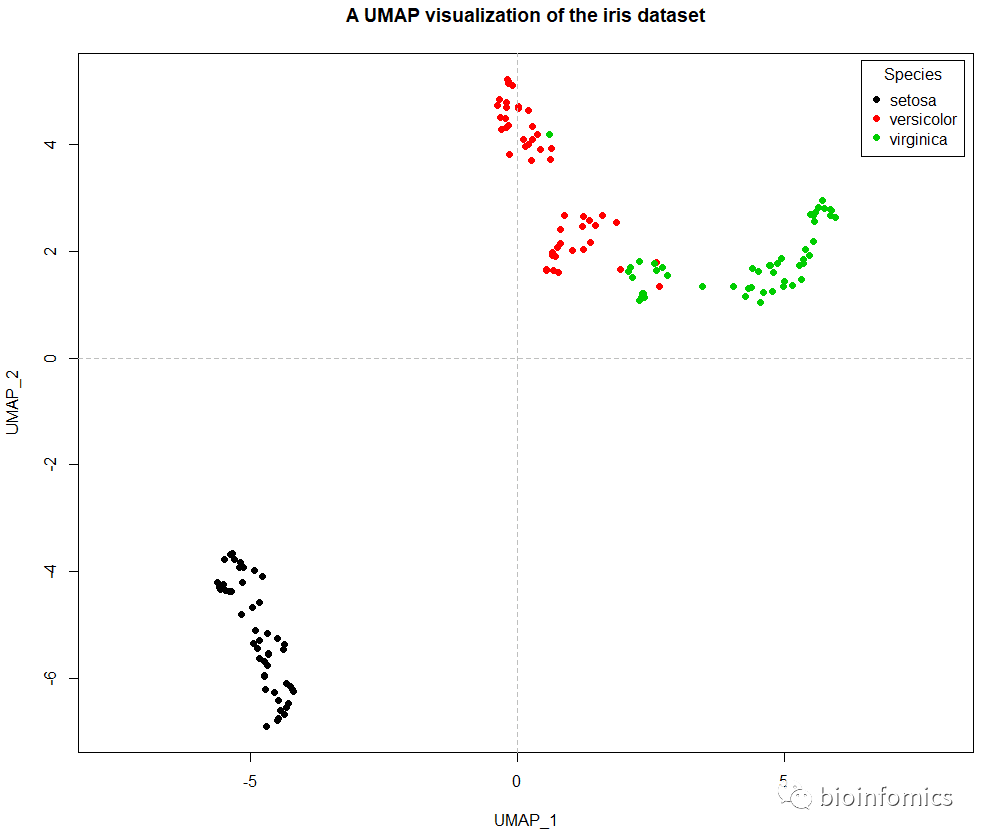
# 使用plot函数可视化UMAP降维的结果
plot(iris_umap,col=iris$Species,pch=16,asp = 1,
xlab = "UMAP_1",ylab = "UMAP_2",
main = "A UMAP visualization of the iris dataset")
# 添加分隔线
abline(h=0,v=0,lty=2,col="gray")
# 添加图例
legend("topright",title = "Species",inset = 0.01,
legend = unique(iris$Species),pch=16,
col = unique(iris$Species))
# Supervised dimension reduction using the 'Species' factor column
iris_sumap <- uwot::umap(iris, n_neighbors = 15, min_dist = 0.001,
y = iris$Species, target_weight = 0.5)
head(iris_sumap)
## [,1] [,2]
## [1,] -10.79467 -14.11691
## [2,] -11.99333 -13.15741
## [3,] -12.39469 -13.71514
## [4,] -12.54123 -13.47225
## [5,] -10.89463 -14.27025
## [6,] -10.15880 -13.58669
iris_sumap_res <- data.frame(iris_sumap,Species=iris$Species)
head(iris_sumap_res)
## X1 X2 Species
## 1 -10.79467 -14.11691 setosa
## 2 -11.99333 -13.15741 setosa
## 3 -12.39469 -13.71514 setosa
## 4 -12.54123 -13.47225 setosa
## 5 -10.89463 -14.27025 setosa
## 6 -10.15880 -13.58669 setosa
# 使用ggplot2包可视化UMAP降维的结果
library(ggplot2)
ggplot(iris_sumap_res,aes(X1,X2,color=Species)) +
geom_point() + theme_bw() +
geom_hline(yintercept = 0,lty=2,col="red") +
geom_vline(xintercept = 0,lty=2,col="blue",lwd=1) +
theme(plot.title = element_text(hjust = 0.5)) +
labs(x="UMAP_1",y="UMAP_2",
title = "A UMAP visualization of the iris dataset")

sessionInfo()
## R version 3.6.0 (2019-04-26)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 18363)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=Chinese (Simplified)_China.936
## [2] LC_CTYPE=Chinese (Simplified)_China.936
## [3] LC_MONETARY=Chinese (Simplified)_China.936
## [4] LC_NUMERIC=C
## [5] LC_TIME=Chinese (Simplified)_China.936
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] ggplot2_3.2.0 uwot_0.1.4 Matrix_1.2-17 umap_0.2.6.0
##
## loaded via a namespace (and not attached):
## [1] Rcpp_1.0.5 RSpectra_0.15-0 compiler_3.6.0
## [4] pillar_1.4.2 tools_3.6.0 digest_0.6.20
## [7] jsonlite_1.6 evaluate_0.14 tibble_2.1.3
## [10] gtable_0.3.0 lattice_0.20-38 pkgconfig_2.0.2
## [13] rlang_0.4.7 yaml_2.2.0 xfun_0.8
## [16] withr_2.1.2 stringr_1.4.0 dplyr_0.8.3
## [19] knitr_1.23 askpass_1.1 tidyselect_0.2.5
## [22] grid_3.6.0 glue_1.3.1 reticulate_1.13
## [25] R6_2.4.0 rmarkdown_1.13 purrr_0.3.2
## [28] magrittr_1.5 scales_1.0.0 htmltools_0.3.6
## [31] assertthat_0.2.1 colorspace_1.4-1 labeling_0.3
## [34] stringi_1.4.3 RcppParallel_4.4.4 openssl_1.4
## [37] lazyeval_0.2.2 munsell_0.5.0 crayon_1.3.4
## [40] FNN_1.1.3

END






文章转载自bioinfomics,如果涉嫌侵权,请发送邮件至:contact@modb.pro进行举报,并提供相关证据,一经查实,墨天轮将立刻删除相关内容。