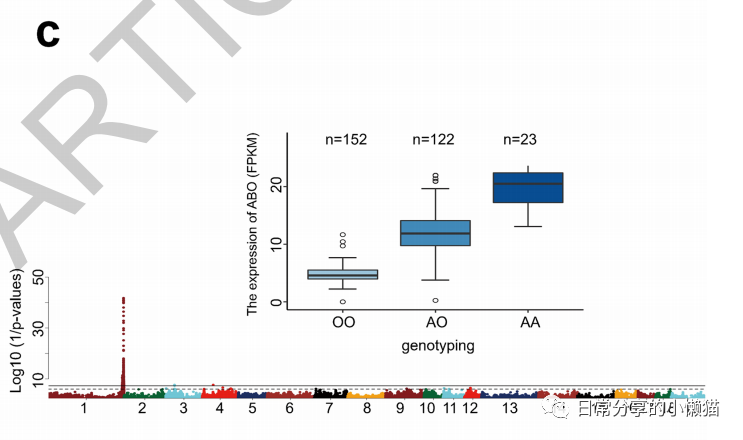
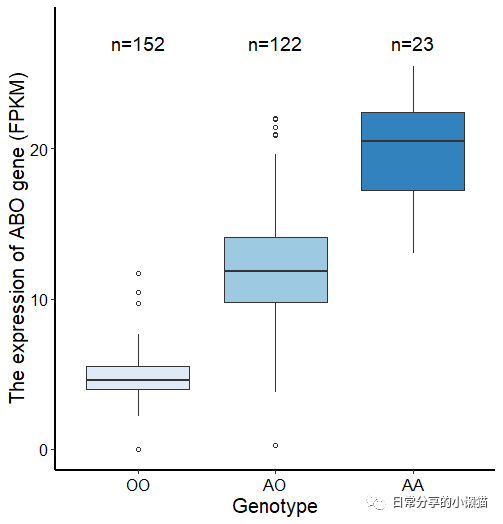
本文主要展示如何利用R语言绘制来自nature期刊ABO genotype alters the gut microbiota by regulating GalNAc levels in pigs(Hui Yang, et al,2022)[1]一文中的Extended Data Fig.5c。该图形式为箱线图,并添加了每组样本的数量。如下图所示:
1、图形绘制
library(ggplot2)
library(EnvStats)
library(dplyr)
ED.fig5 <- read.table("ABO_topSNPExp.txt", header = TRUE)
ED.fig5$Genotype <- factor(ED.fig5$Genotype, levels = ED.fig5$Genotype %>% unique())
head(ED.fig5)
# Genotype GeneExp_UNC
#1 OO NA
#2 OO NA
#3 OO NA
#4 OO NA
#5 AO NA
#6 OO NA
str(ED.fig5)
#'data.frame': 668 obs. of 2 variables:
# $ Genotype : Factor w/ 3 levels "OO","AO","AA": 1 1 1 1 2 1 2 1 2 2 ...
# $ GeneExp_UNC: num NA NA NA NA NA NA NA NA NA NA ...
#绘图
ggplot(ED.fig5, aes( Genotype, GeneExp_UNC, fill = Genotype)) +
geom_boxplot(outlier.shape = 1) +
stat_n_text(size = 5, y.pos = 27) +
scale_y_continuous(limits = c(0,28)) +
theme_classic() +
scale_fill_brewer() +
theme(legend.position = "none",
axis.text = element_text(size = 12, colour = "black"),
axis.title = element_text(size = 15),
axis.line = element_line(size = 1)) +
labs(x = "Genotype",y = "The expression of ABO gene (FPKM)")
 该图绘制较为简洁,关键点在于如何添加样本数量,需要使用到EnvStats包中的stat_n_text() 函数。
该图绘制较为简洁,关键点在于如何添加样本数量,需要使用到EnvStats包中的stat_n_text() 函数。
2、其他
其他绘图方法可进一步阅读公众号其他文章。
如有帮助请多多点赞哦!
参考资料
Yang, H., Wu, J., Huang, X. et al. ABO genotype alters the gut microbiota by regulating GalNAc levels in pigs. Nature 606, 358–367 (2022). : https://doi.org/10.1038/s41586-022-04769-z
文章转载自日常分享的小懒猫,如果涉嫌侵权,请发送邮件至:contact@modb.pro进行举报,并提供相关证据,一经查实,墨天轮将立刻删除相关内容。